New nuclear magnetic resonance study of proteins in nanospace reveals hidden water protein correlations over an enormous range of time scales Dr. Mae-Wan Ho
Proteins are responsible for practically all vital functions, and more and more scientists are coming round to the idea that water plays an essential role in the workings of proteins. But the intimate relationship between proteins and water remains largely hidden because inappropriate methods have been used in studying them, giving an incomplete and even misleading picture. This is very much the message in a report of a new study using a combination of perhaps more appropriate techniques [1].
Researchers at the University of Pennsylvania, Philadelphia, in the United States led by Joshua Wand used reverse micelles, vesicles of surfactants enclosing nano-sized droplets of water dispersed in an organic solvent, to entrap single protein molecules (see Figure 1). In this state, it becomes much easier to probe the relationship between the protein and its hydration water - the water molecules immediately surrounding the protein - using nuclear magnetic resonance (NMR) measurements (see Box).
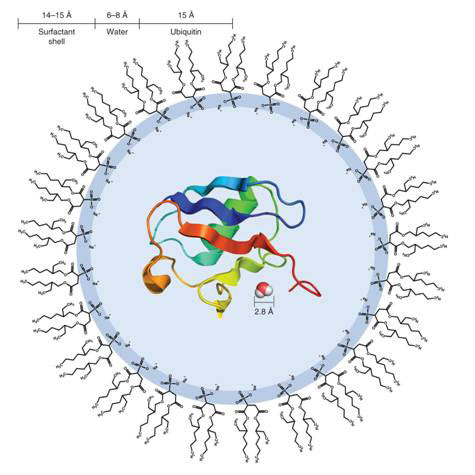
Figure 1 Diagram of a reverse micelle containing a single molecule of ubiquitin
As a result, they found a remarkably diverse range of water molecules associated with the protein molecules that move in concert with different parts of the protein itself, down to individual amino acids, which has never been seen before, overturning a number of previous misconceptions.
The method is ingenious in that it creates conditions much more like the situation inside a living cell, as pioneers of the technique have pointed out (see [2] What's the Bacterium Really Like? SiS 24). Frank Mayer and his team at Gottingen University, Gottingen, Germany, showed how encapsulating enzymes in reversed micelles increased their specific activity 2 to 10 fold, and at the same time greatly improved their thermo-stability [3]. It also increases the precision of nuclear magnetic resonance measurements in focussing on signals from the water immediately around the protein, without being contaminated by the bulk solvent outside, which is an organic solvent, and not water. The confinement in the nano volume has the effect of vastly lengthening the ‘residence time’ of water molecules on the protein (probably by restricting the motion of the protein itself), while decreasing general hydrogen exchange with bulk water, as well as any contribution of long-range coupling to bulk solvent water. This allows the first detailed, site-specific analysis of relative hydration water mobility across an entire protein surface.
The protein used in the study was ubiquitin, a small 8.5 kDa polypeptide of 76 amino acids expressed in all eukaryotic cells (cells with nucleus, in higher organisms). Its main function is in the turnover of proteins [4]. ‘Ubiquitylation’ is an ATP-dependent proteolytic process, in which proteins marked with one or more ubiquitin molecules are rapidly degraded with the release of free ubiquitin.
The reverse micelles were made of the surfactant bis(2-ethylhexyl) sulphosuccinate (AOT), and contains an average of 1 000 molecules including the single protein molecule. The AOT molecules have their hydrophilic (water-loving) acid head groups facing inside the micelle, paired with Na+, rather like membrane lipids in the cell (see Fig. 1). The reorientation of the water confined in the reverse micelle is typically an order of magnitude slower. In order to probe the interaction between protein and water, the protein is heavily labelled with 15N isotope in its amide groups (NH), and the cross-peaks (cross-magnetization, cross-correlation) between the amide H and H of water molecules analysed on 3-dimensional NMR NOE and ROE spectroscopy (see Box).
NMR, NOE and ROE spectroscopy [5-9]
When placed in a magnetic field, NMR (nuclear magnetic resonance) active nuclei of atoms (usually with odd number of nucleons, such as 1H, 13C, and 15N), absorb electromagnetic radiation, and precess (rotate) at a frequency characteristic of the nuclei. A radiofrequency (RF) pulse at 90° to the magnetic field is sent into the sample, and the emission from the sample is monitored. An NMR spectrum is acquired by varying the frequency of the RF radiation, or alternatively, varying the magnetic field. The resonant frequency, and the strength of the signal, is proportional to the strength of the magnetic field. For example, in a 21 tesla magnetic field, protons resonate at 900 MHz; at weaker fields, the resonance frequency would decrease accordingly.
Depending on the local chemical environment, different protons in a molecule resonate at slightly different frequencies. Both this frequency shift and the fundamental resonant frequency are directly proportional to the strength of the magnetic field.
A two dimensional NMR experiment consists of a sequence of two radio frequency (RF) pulses with a delay period in between. It is the timing, frequencies and intensities of these pulses that distinguish one experiment from another. A typical two-dimensional experiment consists of a preparation period where a magnetization coherence is created through a set of RF pulses, p1, followed by the evolution period, t1, when the nuclear spins are allowed to freely precess (rotate), then a second set of pulses, p2, followed by a mixing period where the coherence is manipulated by the pulse to give an observable signal, and the detection period, t2, in which the signal from the sample is registered as a function of time, in a manner identical to one-dimensional NMR. In three dimensional NMR, three RF pulses are used, giving two sets of mixing periods. In four dimensions, four RF pulses are used, with three sets of mixing periods, and so on. Higher dimension NMR is used when large molecules, such as proteins are analyzed, as they are crowded with signals, and spreading them out in more dimensions prevents overlap.
The two dimensions of a 2D NMR are two frequency axes representing a chemical shift. Each frequency axis is associated with one of the two time variables, the length of the evolution period, t1, and the time elapse during the detection period (detection time). These are each converted from a time series to a frequency series by 2-dimensional Fourier transform (a mathematic technique). Thus, a 2-D NMR experiment is generated as a series of one-dimensional experiments by varying the specific evolution time t1. The end result is a 2-dimensional plot showing an intensity value for each pair of frequency variables. For 3-dimensional NMR, the additional dimension is the second mixing period between t1 and t2 and the results are represented as a cube.
A multidimensional NMR spectrum shows two kinds of peaks. Diagonal peaks have the same frequency coordinate on each axis, while cross peaks have different values for each frequency coordinate, and appear off the diagonal. Diagonal peaks correspond to the peaks in a one-dimensional NMR experiment, while the cross peaks indicate couplings between pairs of nuclei, due to magnetization transfer, either between two nuclei in the same molecule (connected by chemical bonds) or else in different molecules nearby, as in the nuclear Overhauser effect (NOE) spectroscopy (NOESY).
Rotating frame nuclear Overhauser effect (ROE) spectroscopy (ROESY) is similar to NOESY except that the initial state is different. Instead of observing cross peaks from an initial state of z magnetization (the usual direction), the equilibrium magnetization is rotated 90° onto the x axis and then spin-locked by an external magnetic field so that it cannot precess.
The NMR spectra of the confined protein in reverse micelles showed dozens of cross peaks representing correlations between specific amino acid amide groups and hydration water. This was in contrast to the relative few peaks identified in ordinary solution NMR of the labelled protein, practically all of which were due to H exchange with nearby labile side-chain hydrogen.
Within the reverse micelles, researchers discovered a diversity of correlated protein-water dynamics, with water rearrangement rates ranging over timescales of more than 1010 from one region of the protein surface to another [10].
The majority of detected hydration sites showed substantial motion or shorter residence time. Approximately three quarters of the amide hydrogens within the NOE detection distance limit (4.3 Å) of solvent showed measurable cross-peaks to water. No cross-peaks to water from the buried amid hydrogens were detected.
A clear group of very slow (residence time ~10 ns or longer) and spatially restricted hydration is evident along the C-terminal tail of the protein, and a cluster of very fast hydration sites is evident along the surface of the a-helix. Clusters of intermediate dynamics are also seen along the mixed b-sheet.
The water molecules in the network of water around the protein seem to act cooperatively locally yet independently of the behaviour of the bulk solvent or of the other regions. Water molecules that have similar hydration dynamics (similar residence times) form clusters across the protein surface.
These results show that the hydrogen-bonded network of water is much more flexible than previously believed, as already hinted at in recent findings on how ions interact with water (see [11] Dancing with Ions, SiS 48).
Until now, much work on site-resolved hydration has relied on crystallographic data. As most protein crystal structures are heavily hydrated, the general view seems to be that protein crystal structures represent the native protein in crowded conditions in the living cell, and hence crystallographic water should also be representative of the native protein in the cell. Extensive analysis of crystallographic water has been combined with molecular dynamic simulation data to build up a picture of the protein hydration layer. Ubiquitin is a case in point.
Two crystal structures for wild-type human ubiquitin have been proposed; only about half of the identified water molecules in the two structures are within 1Å of each other, even though the proteins were crystallized under almost identical conditions. Furthermore, there is little correlation between the waters detected by solution NMR and the locations and occupancies of the crystallographic waters. Notably, only ~6o percent of the crystallographic waters common to both crystal structures appear within the 4.3 Å NOE detection distance of sites where the NMR detected waters are long-lived. Further, more than half of the NMR-detected long-lived protein water interactions involve amide hydrogens that are outside NOE distance from the nearest crystallographic water molecule common to both crystal structures. In addition, there was no correlation between the NOE/ROE values for the NMR-detected sites of long-lived hydration and the location of the nearest crystallographic water common to both crystal structures.
Cell biologists have finally come round to the view that cells are packed with macromolecular surfaces, which confine proteins to nanometre volumes; and confinement can potentially change the fundamental properties of proteins, much as the pioneers Frank Mayer [3] and Jim Clegg [12], for example, have been telling us for years.
Article first published 02/11/11
Comments are now closed for this article
There are 4 comments on this article.
Brad Bartholomew Comment left 3rd November 2011 19:07:13
This seems to have a connection to article I read recently "Light as a Trigger and a Probe of the Internal Dynamics of Living Organsims." This water seems to ramp up the energy from extremely weak biophotons emitted from DNA so that it can initiate cellular processes and bodily functions.
mae-wan ho Comment left 5th November 2011 21:09:57
Brad,
please give the link to the article you came across, and I'll try to give you a considered response.
Ehnriko Comment left 9th November 2011 02:02:43
It's just a matter of time... we are learning more and more about the mystery in water so much recently... I believe it's just a matter of time before we can solve the all Global problems...
Peter Payne Comment left 26th February 2012 22:10:57
Just in case this reference has not reached you--
http://www.mendeley.com/research/light-trigger-probe-internal-dynamics-living-organisms/