A new gene-editing technique has taken the world by storm, it can disable or change specific gene in the genome of all animals including humans faster and more efficiently than ever before but it has raise unprecedented concern over safety and ethics Dr. Mae-Wan Ho
The new gene-editing technique CRISPR (see Box) [1] has taken the world by storm. It enables geneticists to disable or change the sequence of specific genes in the genome of practically all animals including humans faster, more efficiently than ever before, promising to improve our understanding of how genes work, delete genes that cause diseases, even modify human embryos to rid them of diseases or to ‘enhance ‘ them. The applications are moving ahead so fast that many scientists are calling for caution as major safety and ethical concerns need to be addressed.
The issue came to a head when a team of Chinese researchers created the first genetically modified human embryos using the technology.
CRISPR (clustered regularly interspaced short palindromic repeat) refer to short, partially repeated DNA found in the genome of bacteria and other microorganisms that protect the organism against viruses (see Figure 1).
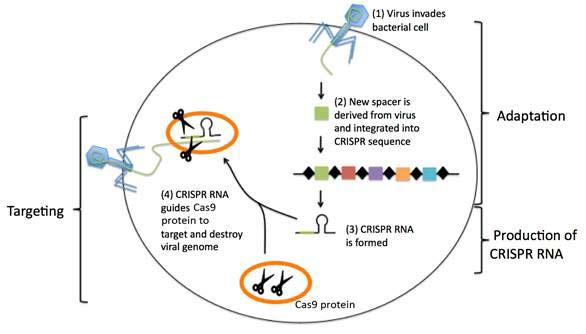
Figure 1 CRISPR-mediated immunity (see text)
In Fig. 1, CRISPR regions are composed of short DNA repeats (black diamonds) and spacers (colour boxes). When a new virus infects the bacterium, a new spacer derived from the virus is incorporated among the existing spacers. The CRISPR sequence is transcribed and processed to generate short CRISPR RNA molecules. The CRISPR RNA associates with and guides bacterial DNA cutting protein (Cas9 protein) to a matching target sequence in the invading virus. The Cas9 protein cuts up and destroys the invading viral genome.
CRISPR has become the latest gene-editing technique, which enables precise changes to be made in the genes of fruit flies, fish, mice, plants and human cells. To do that, geneticist first design and synthesize short a RNA molecule that match a specific DNA sequence. Then, as in the targeting step of the bacterial system, this guide RNA shuttles the Cas9 protein to the intended DNA target, and can silence a gene or change the sequence of a gene by adding a repair template with a specified change in sequence, so that it is incorporated into the DNA during the repair process. The targeted DNA is now altered to carry the new sequence (see Figure 2).
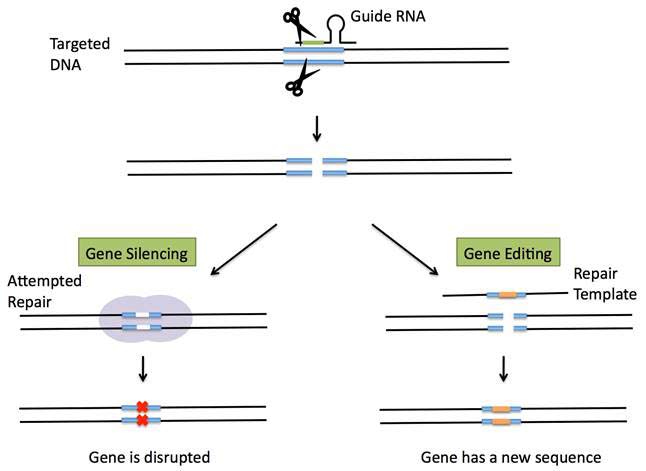
Figure 2 CRISPR gene silencing or gene editing
A team of researchers at Sun Yat-sen University in Guangzhou China used CRISPR to edit the b-haemoglobin gene (HBB) in human pre-implantation embryos [2]. Actually the team used defective embryos with three pronuclei that normally would not be implanted. They found that CRISPR/Cas9 could effectively cut the HBB gene, but the efficiency of homologous recombination directed repair of HBB was low and the edited embryos were mosaic. Off-target cleavage was also found. Moreover, the endogenous d-haemoglobin gene (HBD), which is homologous to HBB, competed with exogenous donor sequence to act as the repair template, resulting in untoward mutations. The researchers concluded: “Taken together, our work highlights the pressing need to further improve the fidelity and specificity of the CRISPR/Cas9 platform, a prerequisite for any clinical applications of CRISPR/Cas9-mediated editing.”
Even before the official publication of the paper, two independent groups of scientists wrote editorials in the journals Nature [3] and Science [4] respectively expressing their concerns. The group in Nature [3] called for a halt to editing the human germ line on grounds that heritable human genetic modifications pose serious risks, and the therapeutic benefits are tenuous. It would also jeopardize current efforts to apply the technology to somatic cells as a potential functional cure for HIV/AIDs and b-thalassaemia. Studies using gene-editing in animals such as rats, cattle, sheep and pigs show that it is possible to delete or disable genes in an embryo – a simpler process than actually correcting or changing the DNA sequence – in only some of the cells. The precise effects of genetic modification of an embryo may not be known until after birth, and long after birth. Patient safety is the primary concern, before ethical concerns are considered. Today some 40 countries ban it. Fifteen of 22 nations in Europe prohibit the modification of the germ line. The US NIH’s Recombinant DNA Advisory Committee explicitly states that it “will not at present entertain proposals for germ line alterations.” Non-therapeutic genetic enhancement is also a concern.
The group in Science [4] emphasize that research is needed to understand and manage risks arising from the use of the CRISPR-Cas9 technology. Considerations include the possibility of off-target alterations, as well as on-target events that have unintended effects. It is critical to implement standardized benchmarking methods to determine the frequency of off-target effects and to assess the physiology of cells and tissues that have undergone genome editing. The potential safety and efficacy issues arising from the use of this technology must be thoroughly investigated and understood before any attempts at human engineering are sanctioned.
Just before an International summit in December 2015 co-sponsored by the US National Academy of Sciences, the US National Academy of Medicine, the UK Royal Society and the Chinese Academy of Sciences to consider the scientific and social implications of genome editing, Jennifer Doudna, a researcher at University of California Berkeley who helped invent CRISPR/Cas9, and a signatory on the editorial in Science, wrote in a commentary in Nature [5] stating that “we do not yet know enough about the capabilities and limits of the new technologies, especially when it comes to creating heritable mutations.” Hence, “human-germline editing for the purposes of creating genome-modified humans should not proceed at this time, partly because of the unknown social consequences, but also because the technology and our knowledge of the human genome are simply not ready to do so safely.” She also stated that future discussions should address other potentially harmful applications of genome editing in non-human system, such as “the alteration of insect DNA to ‘drive’ certain genes into a population” (see below).
In the event, the international summit called for a slowdown on research involving heritable modifications of the human genome [6]. Although the academies acknowledged that such research has the potential to eradicate genetic diseases or enhance human capabilities, they also said the science is just too new to do any of that safely or successfully.
However, they argued it is alright to use germline cells or early human embryos in basic and preclinical lab research, so long as they are not then used “to establish a pregnancy”. (This goes beyond what Francis Collins, National Institutes of Health's director, said on the publication of the China experiment, that the NIH would not fund genomic editing involving human embryos, even if the embryos were not used to create a pregnancy.)
Not all scientists are satisfied with the outcome of the summit [7]. Paul Knoepfler a cell biologist at University of California Davis said he was disappointed that the organisers did not propose at least a temporary moratorium on germline human genetic modification.”
In January 2016, a team led by Keith Joung at Harvard Medical School, Boston, Massachusetts in the United States reported the successful construction of a high-fidelity CRISPR/Cas9 nuclease that has little or no detectable genome-wide off-target effects. This is achieved by changing 4 amino acids with long side chains to one with a short side chain (alanine) to reduce the protein’s non-specific interaction with the phosphate DNA backbone. The resultant SpCas9-HF1 (Streptomyces pyogenes Cas9-high-fidelity 1) nuclease retains on-target activities comparable to the wild type enzyme with 85 % of the single guide-RNAS (sg-RNA) tested in human cells, with very little or no off-target effects. The specificity of SpCas9-HF1 and its potential target was improved with further substitutions to reduce non-specific interactions with the DNA [8].
Reducing off-target effects does contribute substantially to safety. But other safety aspects such as unintended consequences from on-target events and the stability of the modifications, as well as ethical concerns still need to be addressed.
In addition, other worrying applications raise new issues on safety, such as ‘gene drive’ in transgenic insect disease vectors.
The World Health Organization reports that mortality from malaria continues to decrease and estimates that ~3.3 million lives have been saved since 2001 as a result of new drugs, personal protection, environmental modification and other measures; but there were still ~580 000 deaths globally from malaria in 2014 [9].
Researchers at Universities of California Irvine and San Diego has just created a transgenic mosquito in the malaria vector species Anopheles stephensi carrying genes against the malarial pathogen Plasmodium falciparum using CRISPR/Cas9 in a new construct to produce a ‘gene drive’ that makes almost all the progeny of the transgenic male mosquitoes anti-Plasmodium [10].
The ‘gene-drive’ is actually a mutagenic chain reaction first devised by two of the researchers at UC Irvine in Drosophila melanogaster [11]. Previous gene editing was done with Cas9 and the guide RNA and transgene sequence on separate plasmids. By engineering Cas9 next to the gRNA and transgene sequences in a single plasmid, a mutagenic chain reaction is triggered that converts the unmodified gene in the wild-type chromosome to mutant state.
Anopheles stephensi is estimated to be responsible for ~12 % of all transmission in India, mostly in urban environments, accounting for ~106 000 clinical cases in 2014 [3]. Laboratory strains are transformed efficiently with transposable elements to facilitate analysis of transgene expression in diverse genomic locations. Site-specific integration allows insertion of exogenous DNA into the mosquito genome at locations with minimum impact on fitness [12]. Furthermore, a dual anti-parasite gene was developed based on the single-chain antibodies miC3 and m2A10 that target the Plasmodium ookinete protein Chitinase 1 and the cricumsporozoite protein respectively [13]. Transgenic female mosquitoes expressing miC3 and m2A10 were free from P. falciparum sporozoites (infectious stage of the parasite) in their salivary glands under infection conditions expected in the field, and hence incapable of transmitting the parasite. Modeling of gene drive system, which exceed Mendelian inheritance, results in a more rapid transformation of a population with fewer releases than the ‘inundative’ approach, in which engineered mosquitoes (without gene drive) ware released in numbers substantially exceeding those of the local populations [14].
All these considerations encouraged the researchers to construct their gene-drive system in A. stephensi using CRISPR/Cas9-mediated homology-directed repair (HDR) adapted from the mutagenic chain reaction developed in the fruit fly. The drive system as designed works in both the male and female germ lines of mosquitoes derived from transgenic males [10]. Cas9-mediated gene targeting is also evident in the somatic cells of embryos derived from transgenic females. The system can carry a relatively large set of genes (~17kb in length).
The structure of the gene-drive plasmid consists of the Cas9 gene and its promoter, the gRNA and its promoter, the dual antibodies genes with their promoters and marker gene encoding the Drosophila red fluorescent protein (DsRed) with its promoter; this long continuous stretch flanked by sequences homologous to the target the autosomal gene kh encoding the enzyme kynurenine hydroxylase. Altogether, the plasmid is 21 kb in length with 16 625 bp comprising the ‘cargo’ to be inserted into the target gene.
A total of 680 G0 wild-type embryos were injected with the transforming plasmid and other components to aid transformation; 122 and 129 males and females respectively survived to the adult stage, and were assigned to 22 male-founder and 9 female-founder pools and outcrossed to wild type adults of the opposite sex. Two males positive for DsRed, designated 10.1 and 10.2 were recovered after screening 25 723 G1 larvae.
When outcrossed to wild-type females, 10.1 produced all DsRed+ adult progeny (n=14), whereas 10.2 produced only 57 of 129 (44 %) DSRed+ adult progeny. DSRed+G2 males and females from both 10.1 and 10.2 were outcrossed to wild-type mosquitoes, and the G3 larvae progeny were scored for DSRed. Line 10.1 produced 1 321 (99.7 %) DSRed+ while line 10.2 produced 4 631 (99.2 %) DSRed+ G3 larvae. These results are consistent with highly efficient gene-drive.
When the 10.1 and 10.2 G3 males and females were outcrossed to wild type mosquitoes of the opposite sex, the G4 male and female progeny of G3 males and females derived from G2 10.1 and 10.2 transgenic males show a high frequency of DsRed transmission, corresponding to a 96.9 % rate of gene conversion. In contrast, a much high proportion of G4 larvae progeny of G3 males and females derived from G2 10.2 and 10.2 transgenic females appeared to have inherited mutations at the kh locus instead of gene-conversion events with a ratio of 1.33 DsRed+ to 1.0 DsRed-. This indicates that the transgene is unstable in the female, and there as key safety issues involved (see later).
Importantly, the anti-Plasmodium genes are actively transcribed in the DsRed+ mosquitoes.
Long before the transgenic mosquito with gene drive was constructed, a group of scientists have expressed their concerns as the ‘regulatory gap’ regarding such insects [15]. They highlighted the need for containment measures and ‘reverse drive’ to undo the process. They point out that gene drive has not been evaluated for safety. A drive may move through only part of a population before a mutation inactivates the engineered trait. (This was indeed the case in transgenic female mosquito progeny, where the CRISPR/Cas is also active in somatic cells, see above). In some cases preferred phenotypes might be maintained as long as new drive encoding updates are periodically released.
The commentary also made the key point that (p. 627): “In theory, precision drives could limit alteration to target populations, but the reliability of these methods in preventing spread to nontarget or related populations will require assessment. To what extent and over what period of time might crossbreeding or lateral [i.e., horizontal] gene transfer allow a drive to move beyond target populations? Might it subsequently evolve to regain drive capabilities in populations not originally targeted?”
This is crucial in the light of the instability of the gene drive in transgenic female mosquitoes reported [10]. When these females bite animals including humans, there is indeed the possibility of horizontal gene transfer of parts, or the entire gene-drive construct, with potentially serious effects on animal and human health. Cas9 nuclease could insert randomly or otherwise into the host genome, causing insertion mutagenesis that could trigger cancer or activate dominant viruses. In addition, the many transcripts and gene products encoded by the gene-drive construct could also have harmful effects including immunological reactions. These same hazards apply, all the more so to species of animals that feed on mosquitoes, as it is now well-known that nucleic acids in food can get into cells and tissues (see [16] Nucleic Acid Invaders from Food Confirmed, SiS 63).
Finally, the ecological risks of gene drives are enormous, so warns conservation scientists from Australia’s Commonwealth Scientific and Industrial Research Organisation [17]. They stated: “The question is no longer whether we can control invasive species using gene drive, but whether we should.” As the gene drive can in principle lead to the extinction of a species, this could involve the species in its native habitat as well as where it is considered invasive. As distinct from conventional biological control, which can be applied locally, there is no way to control gene flow. They point out that because the CRISPR/Cas gene drive remains fully functional in the mutated strain after it is created, the chance of off-target mutations also remain and the likelihood increases with every generation. “If there is any risk of gene flow between the target species and other species, then there is also a risk that the modified sequence could be transferred and the adverse trait manifested in nontarget organisms.” (This commentary has not even begun to consider horizontal gene flow, which would multiply the risks many-fold.)
There is also increasing awareness that many invasive species will have considerable niche overlap, such that removal of one species will enable another to rapidly take its place.
They call for a thorough ecological risk assessment before any application of CRISPR/Cas gene drive is contemplated in the control of alien species, to prevent a ‘silver bullet’ becoming a ‘conservation threat’.
There are numerous reasons to proceed with caution with CRISPR/Cas9 applications. It is a powerful, efficient, and cheap gene-editing tool beset with risks for health and the environment. Particularly worrying is its use in gene-drive, a currently irreversible and uncontrollable process once released into the environment.
Article first published 12/01/16
Comments are now closed for this article
There are 2 comments on this article.
Todd Millions Comment left 14th January 2016 01:01:53
I'm confused! According to the our Sages of P.R. science-we already have such exquisite precision in our gene engineering.With NO off target effects and utter stability and predictability!This MUST be true as all our holy bureaucrats and sacred administrators SAY so(except for the heretics). So why do we need a new one? Has something gone wrong?
Rory Short Comment left 15th January 2016 23:11:37
This is exciting new technology but it is new therefore we should proceed with the utmost caution and keep it firmly in the lab for now.