Geneticists can now create millions of new viruses and bacteria in a matter of minutes in the laboratory. And there is no regulation to stop them from being effectively released. Dr. Mae-Wan Ho reports.
Willem P.C. Stemmer of Affymax Research Institute in Palo Alto, California, published a paper in the Proceedings of the National Academy Science in 1994, describing a technique for rapidly shuffling and recombining DNA in the laboratory that appears to generate recombinant proteins with greatly improved performances.
According to Stemmer, computer simulation studies had demonstrated the importance of recombining segments of genes in evolution, rather than single base changes. To put this into practice, he devised a method for reassembling genes from random gene fragments in the test tube.
The DNA of the gene is digested with the enzyme deoxyribonuclease 1 (DNAse 1) into random 10 to 50 bp (base-pair) fragments. These fragments are heated up to separate the two strands and made to re-anneal (pair up again) in the presence of a DNA polymerase, an enzyme that synthesize DNA.
The separated strands of DNA pair up according to complementary base sequences in homologous DNA (DNA with similar base sequences from the same species or related species); and a shorter sequence paired to a longer one will 'prime' the synthesis of DNA using the longer sequence as a template, until the complete double stranded DNA is restored. With fragments re-annealing in a random way, the DNA polymerase can switch templates many times in the course of reassembly, and that is how different recombinants are generated. A simplified diagram of what is achieved in a typical experiment is shown in Figure 1.
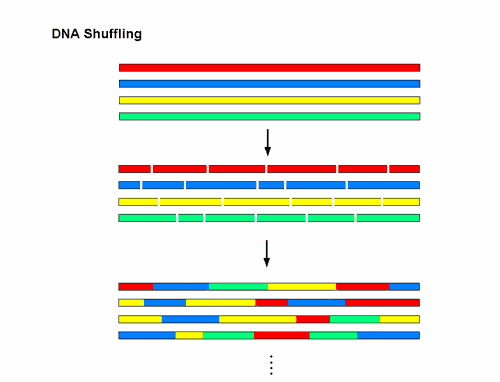
Figure 1 DNA shuffling. Different variants of a gene or a DNA sequence, represented by the different colours are fragmented and reassembled to generate millions of recombinants all at once.
The recombinants are cloned into a vector and introduced into bacteria for rapid screening.
In this way, many variants of any gene can be rapidly recombined, even variants from different species. Larger pieces of DNA such as entire plasmids, viral and bacterial genomes can also be recombined. To increase the variety of recombinants, short pieces of synthetic DNA can be added into the mixture. Even non-homologous DNA, from totally unrelated species can be recombined.
In the very first experiment reported, Stemmer claimed to have produced a recombinant of a beta-lactamase that increased resistance to a beta-lactam antibiotic by 16 000 fold. When this recombinant is 'backcrossed' to the parental gene using the same DNA shuffling technique, which he later calls, 'molecular breeding', an enzyme that was 32 000 times as effective as the wild type enzyme was obtained.
Previously, researchers have used other methods to introduce random errors, such as the polymerase chain reaction (PCR), but that gave only a 16 fold improvement at best. And 'rational design' based on known structure of the enzyme did no better. So the results obtained with DNA shuffling seem out of this world.
Stemmer has since set up a California-based company, Maxygen, Inc. to exploit the technology. In a series of papers between 1998 and 2002, he and his coworkers in Maxygen and other companies have used the technique to obtain greatly improved proteins, such as green fluorescent protein, fucosidase, subtilisin, thymidine kinase and interferon-alpha, as well as improved retroviral vectors and bacterial strains of Streptomyces and Lactobacillus.
In the experiment on retroviral genomes, 6 mouse leukaemia viruses (MLV) were recombined in a single round, giving a library of 5 million replicating recombinant viruses. Among them were completely new viruses that infect Chinese Hamster Ovary cells, which none of the original MLV was capable of infecting, and recombinant viruses that were 30 to 100 times more stable than the parental strains, and hence much better for gene therapy.
So far, such experiments appear to have been carried out exclusively by Maxygen Inc. or by the company in collaboration with other companies, and therefore not independently replicated as far as the remarkable successes are concerned.
There is no doubt, however, that the procedure itself is dangerous. As these researchers have amply demonstrated, recombination is the major route to generating new viruses and bacteria. Some of the recombinant viruses and bacteria could well be lethal.
What precautions have they taken with regard to the recombinants and vectors used, to prevent them from being released into the environment? Are the recombinant viruses and bacteria, or their DNA, strictly contained within the laboratory?
Current regulation on contained use of genetically engineered micro-organism is highly inadequate, and we have repeatedly pointed this out to our regulators. In Europe, 'tolerated releases' are allowed, and this include not only transgenic wastes consisting of dead bacteria and cells (with plenty of transgenic DNA), but also certain living bacteria deemed, by the companies concerned, not to pose a threat to health or the environment.
With unregulated experiments like these, who need bio-terrorists?
Article first published 17/04/03
Comments are now closed for this article