New findings in genetics show that evolution happens by precisely targeted natural genetic engineering and not by the natural selection of random mutations, says leading molecular biologist James Shapiro, but what are the implications for the safety of GMOs and social policies? Dr. Mae-Wan Ho
I have been awaiting his latest papers for years ever since he first introduced the concept of ‘natural genetic engineering’ in 1997 [1], referring to organisms themselves using ‘cut and splice’ techniques to meet environmental challenges, same as those used by human genetic engineers in the lab. It was a major inspiration for my book [2] Genetic Engineering: Dream or Nightmare? (ISIS publication) warning of dangers from genetically modified organisms (GMOs) released into the environment.
James Shapiro at University of Chicago Illinois in the United States is among the pioneers who discovered the new genetics of the ‘fluid genome’ that, by the early 1980s, had already shaken the scientific establishment to its roots [3] (see also [2, 4], [5] Living with the Fluid Genome, ISIS publication). All the basic tenets of conventional genetics that had dominated science and society for at least half a century were being eroded by exceptions upon exceptions, until the exceptions outnumbered and overwhelmed the rules.
In his 1997 paper [1], Shapiro made a powerful case against the neo-Darwinian dogma that evolution occurs by the natural selection of random mutations. Bacterial genomes typically have a modular structure consisting of sets of genes (operons) expressed together. Operons have a characteristic internal structure, with coding regions for regulator protein(s), structural/enzyme proteins and several control elements. Every protein coding sequence in turn contains several domains each with a defined function. Many operons are dispersed throughout the genome, equipped with different combinations of similar regulatory/control genetic elements and forming overlapping, often extensive ‘regulons’ (gene expression networks) finely tuned to different stages of cell growth and development and changing environmental conditions. Staggering multitudes of protein-effector, protein-protein, protein-nucleic acid, and nucleic acid-nucleic acid interactions are involved, all highly specific for every occasion. Not surprisingly, genomes have special mechanisms for correcting base sequence errors during DNA replication.
It is extremely hard to imagine how such a genome could have been assembled or changed piece-meal by the natural selection of independently occurring random mutations in different genetic elements. On the other hand, a simple copying (amplification) process followed by cut and splice different sequence elements together would be easily accomplished. Cells have all the enzymes and cofactors required for such feats of natural genetic engineering. In fact, artificial genetic engineering is possible only by using the enzymes isolated from the bacteria themselves, albeit without the precision and finesse of natural genetic engineering.
Shapiro discovered ‘adaptive’ mutations in bacteria (recently confirmed, see [6] Non-Random Directed Mutations Confirmed, SiS 60). He investigated an E. coli system that depends on generating a fusion lacZ protein, the b-galactosidase that breaks down lactose to galactose and glucose. The bacterial virus (phage) Mu was used to construct a strain in which a defective lacZ coding sequence without its promoter - a control element required for transcription - and carrying an ochre triplet (a stop codon) at codon 17 - so the transcript cannot be translated fully - was aligned in tandem with another coding sequence araB (from the arabinose operon) that has an intact promoter [1]. In that way, a precise deletion of intervening sequence is needed to form the fusion b-galactosidase protein capable of functioning to break down lactose and enable the cell to growth on a selective medium with lactose as the sole carbon source.
Shapiro originally thought that the Mu prophage (phage integrated into the bacterial genome) would be the passive source of homology (sequence similarity) to enable the fusion to take place by homologous recombination to loop out the intervening sequence, and such ‘spontaneous’ break-rejoin events would generate the actual fusions by removing all blocks to transcription and translation between araB and a site in lacZ downstream of the ochre triplet codon. But detailed studies showed that the Mu prophage played an active role in the araB-LacZ fusions using its transposase enzyme, and the process was precisely regulated by the cell. Many different proteins and DNA sequences have to come together in choreographed succession to form and rearrange the nucleoprotein complexes necessary for directing the precise cut and splice operations. A large number of the molecular players have been identified since. In other words, the fusion events happen as the result of accurate natural genetic engineering carried out by the E. coli cell.
As mobile genetic elements like Mu are found in all organisms, Shapiro thought it reasonable to hypothesize that the regulatory aspects of the mutational process exemplified by the araB-LacZ system might apply generally to other examples of adaptive mutations (see [6]) and described the numerous cellular functions involved in different cases. He wrote [1, p.103]: “The depth of regulatory interactions between cellular signal transduction networks and natural genetic engineering systems is likely to prove typical rather than exception.”
Natural genetic engineering has large implications for evolution, Shapiro pointed out. First, large scale coordinated changes within the genomes of single cells are possible because a particular natural genetic engineering system can be activated to operate at multiple sites in the genome. Second, there is opportunity for adaptive feedback to make genetic changes, thereby greatly accelerating evolutionary change during episodes of crisis.
In his new papers, Shapiro draws an illuminating parallel between the genome and the computer [7, 8]; at the same time correcting some widely held misconceptions about the genome.
“The genome has traditionally been treated as a Read-Only Memory (ROM) subject to change by copying errors and accidents.” Shapiro writes [7, p. 268]: “I propose that we need to change that perspective and understand the genome as an intricately formatted Read-Write (RW) data storage system constantly subject to cellular modifications and inscriptions.”
The ROM view of the genome is encapsulated by Sydney Brenner in his 2012 Alan Turing Centennial tribute [9]: “Turing’s ideas were carried further in the 1940s by mathematician and engineer John von Neumann, who conceived of a ‘constructor’ machine capable of assembling another according to a description. A universal constructor with its own description would build a machine like itself. To complete the task, the universal constructor needs to copy its description and insert the copy into the offspring machine. Von Neumann noted that if the copying machine made errors, these ‘mutations’ would provide inheritable changes to the progeny.”
This static mechanical view of the genome is a far cry from reality. Even to reproduce a single protein – originally conceptualised as a single message – requires elaborate cut and splice operations. The international research consortium project ENCODE (Encyclopedia of DNA Elements) data have revealed that the vast majority of genomic DNA include many ‘non-coding’ segments [10, 11]. The term ‘gene’, a theoretical construct that has never been possible to define rigorously, is now known to be scattered in bits across the genome, overlapping with bits of multiple genes that have to be spliced together to make a messenger (m)RNA for translation into protein. The term now used for the bits is ‘coding sequences’ or exons.
The Turing tape analogy does not take into account the actual physical participation of the genome in productive and regulatory interactions. The concept of a Read-Only Turing genome also fails to recognize the essential ‘Write’ capability of a universal Turing machine, which fits remarkably well with the ability of cells to make temporary or permanent inscriptions in DNA.
(Of course, it is by no means all down to the genome. A genome outside a cell can do nothing. The numerous claims that synthetic biologists have created life in the laboratory are spurious, as they all depend on putting a synthetic genome into a pre-existing cell [12] (Synthetic Life? Not By a Long Shot, SiS 47). Moreover, it is not so much the cell, but rather the nature of living protoplasm that keeps eluding our grasp [13, 14] The Rainbow and the Worm, The Physics of Organisms, and Living Rainbow H2O, ISIS publications.)
Shapiro [1, 7, 8] distinguishes modifications of DNA (rearrangements, deletions, insertions, mutations) - which he regards as natural genetic engineering proper - from epigenetic changes involving DNA/histone marks, or via non-coding RNA species that occur constantly in real time within the life cycle of the cell or organism. In my view, this distinction is artificial. There is no real separation between epigenetic and genetic; they form one seamless continuum in molecular mechanisms that interact with one another directly. In a further paper [15], Shapiro himself proposes that during ‘life history events’ such as hybridization and chromosome doubling, viral or bacterial infections, exposure to environmental toxins, etc., epigenetic changes are often accompanied by mobilization of transposable elements that change the genome. And non-coding RNAs (ncRNAs) are involved in mobilizing transposons and in targeting specific changes in chromatin, the DNA histone protein complex that forms a chromosome. Another common connection between epigenetic and genome change is that processed, alternatively spliced RNA can be reversed transcribed and inserted into the genome. On the other hand, certain altered (reformatted) states of /chromatin can be passed on to subsequent generations; i.e., they are inherited like a mutation. And various species of interference RNA can also act independently as genetic material to perpetrate epigenetic changes across many generations, as part and parcel of the hereditary legacy of the organism (see [16] RNA Inheritance of Acquired Characters, SiS 63). In the new genetics of the ‘fluid genome’, the genome is no longer the constant and unchanging entity previously assumed. Hence I use the term “natural genetic modification” for the totality of changes in the genetic information of cells and organisms as they experience their environments that are all necessary for survival, and some of which are passed on to the next generation(s) [17].
We shall follow Shapiro’s story [7, 8] on actual modifications of DNA base sequence and the genome structure before dealing with implications on artificial genetic modification and for society in general. The rest of this series of articles will elaborate on the epigenetic aspects of natural genetic modification.
One of the most intriguing features of genomic DNA is that it is highly redundant. Many DNA elements are repeated in the genome. Some are grouped together as tandem repeats or short satellites, while others are dispersed at different sites. The repeats arose from active amplification processes. In the human genome, recent survey indicates that up to 67 % is repetitive DNA, while the protein-coding exons amount to less than 2 %.
Much of the genome is composed of defined DNA elements that are complex data cassettes, comprising coding sequences, transcription signals, splice sites and other classes of functionally significant sequences. Regardless of where they insert into the genome, these cassettes will have significant and predictable effects on the functioning of nearby DNA sequences.
Shortly after repetitive DNA was discovered as an abundant genome component in the late 1960s, Roy Britten (1919-2012) and Eric Davidson (now at Caltech in the United States) proposed that repetitive elements could constitute distributed regulatory sites and form networks linking distant genetic loci [18]; for example, through regulatory proteins or RNA molecules that bind to those sites to turn the genes on or off. This has been amply confirmed by discoveries since, in both prokaryotes and eukaryotes.
Genome sizes of organisms range over 6 orders of magnitude (~0.3-100 000 Mbp), but all are highly organised. For circular genomes (in most bacteria), it is necessary to specify the origins of replication, and where it ends to make sure both strands are fully replicated. For linear DNA molecules in eukaryote (organisms with nucleus) genomes and some bacterial (prokaryote) genomes, special structures are needed to protect the ends to ensure their stability.
Once genomic DNA molecules have been replicated and the two copies separated, they must be moved, each to one of the two daughter cells at division. This is accomplished by attaching to microtubules in eukaryotes or to tubulin- and actin-like molecules in bacteria. In eukaryotes, microtubules attachment occurs at complex structures called kinetochores, which form at a special chromosome region called the centromere. The centromeres of most eukaryotic chromosomes have particular genetic and chromatin configurations. Typically, centromeres are made of one or more families of tandem repeat elements, and centromeric nucleosomes (see below) contain special histone protein variants.
There are recognitions sites for RNA polymerase (called promoters) and transcription factors for transcriptional formatting; and that’s where a lot of regulation occurs.
Eukaryotic genomes have additional levels of organization inside the nucleus compared with prokaryotes (that have no nucleus), thus providing further layers of regulation and possibilities for natural genetic modification. Eukaryotic genomic DNA is compacted and packaged into complex structures, chromosomes. The first level of compaction is the winding of ~147 base-pairs of DNA around a core of 8 positive charged histone protein molecules to form a nucleosome. There is a code determining where nucleosomes are to be positioned. The nucleosomes form at different densities along the DNA, which then folds into higher order chromatin complexes that may involve additional proteins and RNA molecules.
‘Chromatin’ derives from cytogenetics where chemical stains were used to make chromosomes visible. Lightly stained ‘euchromatin’ has a lower density of nucleosomes and considered to contain active DNA. More intensely staining heterochromatin regions have a higher density of nucleosomes and considered inactive for replication and transcription. Regions of active euchromatin, silent heterochromatin, and other distinct types of chromatin can be separated from each other by DNA elements known as insulators.
The nucleus is highly organized and sub-compartmentalized. There are special foci where discrete genome functions take place, such as replication, transcription, and repair. Other signals called S/MARs (surface/matrix attachment regions) are sites for localization to the nuclear lamina, a dense fibrillar network inside the nucleus composed of intermediate filaments and membrane associated proteins.
LINE elements (long interspersed element), one class of dispersed repetitive DNA that comprises about 20 % of the human genome, have twice the average density of s/MAR sequences. As we shall see, these and other repetitive elements play a major role in crafting the functional architecture of the genome.
Shapiro’s strong message is that natural genetic engineering is specifically activated, targeted and precisely executed. His personal experience with physiological activation of natural genetic engineering led to his discovery of the mobile genetic element mediated fusion of araB and lacZ coding sequences to create the hybrid protein needed for survival (see above). Fusions were not detectable in 3 x 1010 plated bacteria following normal growth, i.e., no colony within the first 3-4 days, but after several days of additional incubation, colonies began to sprout in ever greater numbers and the frequency increased rapidly to at least one fusion per 10% viable cells on the selection plates. The key parameter inducing this particular DNA restructuring was aerobic starvation. The araB-lacZ fusion system was the first example of a more general phenomenon that came to be called ‘adaptive mutation’ or ‘directed mutation’.
There are other examples. In bacteria, pheromones and quorum sensing signals stimulate the transfer of conjugative elements (during bacterial mating) and uptake of extracellular DNA. In budding yeast, retrotransposition of the Ty3 and Ty5 elements is coordinated with mating events by sex pheromone induction. In rodents, steroid hormones induce reproduction and retrotransposition. In the mouse and human immune systems, lymphokine molecules determine the sites of DS breaks and NHEJ for isotype switching (see later).
The DNA changes associated with antigen receptor synthesis in lymphocytes are the most thoroughly documented case of a tissue-specific natural genetic engineer. But there are other cases, showing that the immune system is not exceptional. One of the most intriguing tissue-specific regulatory phenomena is the activation of long interspersed elements (LINEs) retrotransposition in mammalian nerve cells. This led to the hypothesis that LINE-induced genome diversity is a contributing factor to neural network architecture in the mammalian nervous system.
In principle, homologous recombination can occur between any DNA duplexes that have the same sequence; but actual in vivo recombination displays hot and cold spots, and has all the signs of being tightly regulated. An example of sequence specificity is provided by the chi sites, short bacterial genome sequences near which homologous recombination is more likely than expected. They regulate motor-protein and exonuclease specificity of proteins needed to process broken duplex ends to initiate bacteria recombination. This feature of the prokaryotic recombination process has evolved independently at least twice, given that gram negative and gram-positive bacteria have distinct sets of chi sites with matching exonuclease.
An example of targeting homologous exchange away from particular regions of the genome comes from studies in mice, where recombination events might disrupt working combinations of transcriptional regulatory signals. A particular protein has evolved to recognize those combinations and to suppress them. This same protein also protects the integrity of its own coding sequences.
Another example of targeted natural genetic engineering is that mobile elements are directed towards adaptively useful locations and away from harmful regions, though protein-DNA and protein-protein interactions. For example, the bacterial transposon Tn7 encoding antibiotic resistances when horizontally transferred into a new cell is integrated at a specific locus on the bacterial chromosome by a DNA-recognition multiprotein integration complex so that it has no deleterious effect on bacterial physiology. However, when a plasmid without Tn7 is leaving a cell that carries a chromosomal copy of the element, a different targeting protein becomes part of the transposition/integrating complex. That particular protein binds to a DNA replication factor and targets new insertions in a sequence-independent manner to actively replicating DNA. As replication in the donor cell powers plasmid transfer events, this protein-protein interaction targets Tn7 to mobilizing plasmids and facilitates dispersal of the transposon to new cells. In yeast, there are retroviral-like mobile elements that display distinctive insertion patterns determined by protein-protein interactions that keep them from disrupting expressed coding sequences and allow them to alter coding sequence expression.
We shall look at a number of different targeted natural genetic engineering mechanisms.
Cells are responsible for maintaining the stability of the genome during cycles of growth and reproduction as well as during stationary differentiated states. On the other hand, they can also execute precise genome changes under different environmental challenges. Most certainly, cells and their genomes are not [7] “passive victims of replication errors or DNA damage.”
Exonuclease proof-reading during DNA replication, and mismatch repair afterwards catch 99.9 to 99.99 % of errors. Under optimal conditions, an E. coli cell replicates its DNA at the rate of some 2 000 bp per second. The DNA polymerase has an intrinsic error frequency of about 10-5. Within two proof-reading steps, this is reduced to less than 10-9. This amazing precision results from sensing functions and the recruitment of correction mechanisms.
However, there are both error-free repair processes and error-prone mutator processes in cells. Apoptosis (programmed cell death) is another way of avoiding irreparable DNA damage; this option is present in all types of cells if DNA damage cannot be corrected or bypassed. Cells are constantly evaluating both intracellular and extracellular information to decide on the appropriate action.
There are apoptosis-promoting intercellular signals in both prokaryotes and eukaryotes (see [8]). In mammalian cell culture, the presence of growth factors such as insulin-like grow factor (IGF) protects against apoptosis and leads to DNA repair, while the presence of death factors such as tumour necrosis factor (TNF) induces apoptosis,
For broken DNA duplexes, repair by homologous recombination (HR) with an undamaged template is intrinsically error free. NHEJ in contrast creates new sequence structures, and is therefore inherently prone to error and mutagenic. The distinction is less clear in other cases. For example, base excision repair that removes uracil from DNA by uracil-n-glycosylase (Ung) can be either accurate repair as in E. coli or mutagenic as in activated B human lymphocytes. In the immune system, Ung activity is essential to somatic hypermutation for antibody refinement.
UV induced mutagenesis in E. coli was regarded a random process until it was found to be actively controlled by the cell in a ‘SOS’ DNA damage response. The UV-inducible SOS functions DinB and UmuCD play key roles in mutagenesis. DinB encodes polymerase IV and UmuCD encodes DNA polymerase V, both typical of a class of polymerases that can elongate newly synthesized DNA strands a few nucleotides long opposite a damaged template strand (trans-lesion bypass polymerase), and highly prone to errors. Thus, they have been called mutator DNA polymerases. Detailed study of UV mutagenesis in E. coli revealed that particular trans-lesion polymerases or pairs thereof are necessary for specific kinds of mutations, such as -1 or -2 frame-shifts or particular base substitutions. These unexpected findings indicate that individual localized point mutations are carefully crafted.
When DS DNA breaks occur in eukaryotic cells, special chromatin containing exceptional histones H2A (in yeast) or H2AX (in mammals) forms at the broken ends. Using immunofluorescence and fluorescent protein tags, it is possible to see that broken ends, homologous recombination proteins, and NJEJ proteins localize to sub-nuclear repair centres in a cell cycle-dependent manner. By differential labelling of DNA, the presence of broken ends from two different chromosomes can be seen in a single repair centre where the ends can be joined by NHEJ to form a rearranged structure. For recombination and NHEJ to take place, motor proteins are needed for chromatin remodelling of the damaged DNA and powering the DNA movements to repair centres and executing strand exchanges.
In addition to systems used for DNA repair and genome change, there are a number of evolved complex molecular systems dedicated to generating novel genome structures.
Horizontal gene transfer (hgt) by direct uptake of DNA from the environment, virus infection, conjugation (bacterial mating), parasite vectors and other yet unknown means made it possible for genomes to acquire coding capacity for new activities. The importance of hgt in spreading antibiotic resistance and special adaptations among bacteria and archaea is widely recognized. Less well known are a rapidly growing number of examples in which eukaryotic microbes and multicellular eukaryotes have acquired adaptive functions from prokarytoes or other eukaryotes by hgt.
Viruses are agents of gene transfer as well as agents of infectious diseases. They expand the collective read-write genome of living cells. Sorin Sonea in the 1970s and 1980s [19] viewed the prokaryotic genome as an ecology-wide distributed system containing DNA sequences encoding all kinds of adaptive functions. When a particular ecological niche became available, the requisite functions could be assembled in one cell by horizontal DNA transfer, including viral infections to produce an organism that could exploit the new niche. This view is corroborated by results of environmental metagenomics, which show that viruses in totality provide a significant reservoir of DNA encoding a wide range of cellular functions.
The sequenced genomes of the recently discovered giant DNA viruses that infect amoebae, algae and other protists reveal the extent of genome mixing between the three domains of life. The megabase-range genomes of these viruses contain viral, archaeal, bacterial and eukaryotic sequences. Large DNA viruses have extended host ranges and the protists they infect harbour bacterial symbionts that also infect plants and animals. There is even evidence of conjugate transfer within amoebae between animal and plant pathogenic bacteria. Thus, there exist multiple biological paths for distributing novelties originating within the lower eukaryote-large DNA virus melting pot for genome sequence innovation in all kinds of cells. (That is why I consider the horizontal spread of synthetic transgenes released into the environment as a most serious threat to health and the environment [2]).
DNA rearrangements serve important functions. They provide adaptive immunity to cells against infectious microbial agents, and also enable the infectious agents to vary their surface antigens for escaping immune defences. In addition, site-specific recombination ‘shufflons’ expand the specificity of the bacterial pilus (cylindrical filament involved in mating) for attaching to another bacterial cell for plasmid transfer. And ‘diversity-generating retroelements’ extend the host range of bacteriophages.
Class switching is a well-known example of DNA rearrangement in somatic cells. This process occurs in activated B cells already producing immunoglobulin antibodies after an initial elaborate DNA rearrangement that joined together three separate coding sequences (V, D, J) to make the immunoglobulin (Ig) chains and directed hypermutation at the antigen-recognition sites to produce high affinity antibodies. Immunoglobulins originate as cell-bound IgM. To direct the specific immunoglobulin to different locations in the body, a change in the constant region of the heavy chain is required. This is achieved by class switch of the encoding heavy chain to another Ig class or isotype. The switch occurs by inducing DS breaks at special switch regions preceding each heavy chain exon at the immunoglobulin heavy chain IgH locus (see Figure 1). As only activated and transcribed switch regions undergo breaks follows by end-joining, the process requires a specific combination of lymphokines (intercellular signalling molecules) bound to the promoter of the switch region preceding each exon, as well as the NHEJ enzymes. The cells of the immune system effectively instruct the activated B cells which isotype antibody to synthesize. The connection of transcription and DNA rearrangement is a powerful tool for directing natural genetic engineering activities to functionally regulated region of the genome.
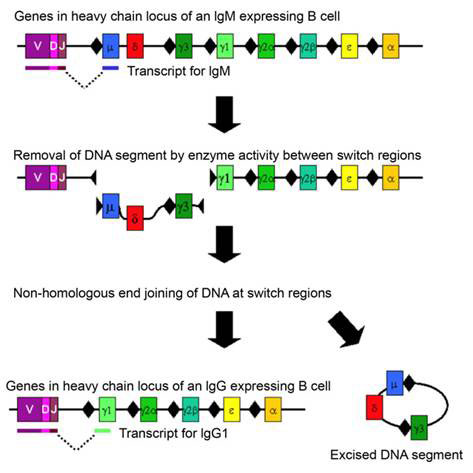
Figure 1 Chromosome rearrangement in class switching of immunoglobulin heavy chain; switch regions are indicated by black diamonds before each exon
A fascinating example of radical genome rearrangement occurs in ciliate protozoa, a large family of eukaryotic microbes that carry both germline and somatic nuclei in the same cell. During vegetative growth by mitotic cell division, the larger somatic macronucleus encodes the RNA and protein molecules needed for reproduction and the small germline micronucleus maintains the complete genome needed for sexual reproduction. Macronuclear somatic chromosomes are small, often carrying a single coding sequence in high copy numbers. Micronuclear germline chromosomes are typical eukaryotic chromosomes and diploid, carrying many sequences absent from the somatic macronucleus. When food is scarce, ciliates undergo meiosis and mating. Afterwards, the old macronucleus degenerates and a new one developed from a descendant of the post-mating zygote nucleus, guided by RNA molecule from the old macronucleus. The process is complex and involves elimination of much germline-specific DNA, up to 90 % depending on the species, and reassembling the scrambled germline sequences into accurate coding sequences. Thousands of DNA cutting and splicing events are needed to put together a working genome within a few hours, and is the most elaborate example known of rapid restructuring of genome architecture within a single cell generation.
Insertion of a mobile genetic element in or near a particular genetic locus is among the commonest ways to alter regulation of an existing genetic locus. The regulatory changes involve transcription factor, micro (mi)RNA and RNA-directed epigenetic controls. New exons frequently arise after insertion by using splice signals inside the mobile elements. Fragments of viral and organelle sequences have also undergone ‘exonization’ to generate novel proteins and domains of proteins in nuclear genomes.
There are numerous examples of transcription factor regulatory sites mobilized by transposons, retrotransposons and retroviruses. Epigenetic imprinting (marking genes to be expressed in a parent-of-origin-specific manner) is often linked to the presence of DNA from mobile elements. Integrated viruses also change the regulatory configuration of the genome. Viruses provide sequences for non-coding (nc)RNAs, sites for transcriptional control, and epigenetic regulation. Genome sequencing has revealed numerous new protein functions arising from the accretion and rearrangement of domains shared by many different proteins.
Mobile genetic elements rapidly disperse common regulatory sequences to distant gene loci to generate coordinately controlled genome networks. Single events can activate mobile genetic element activity that results in multiple genome modifications. This is of particular importance after interspecfic hydridization and whole genome duplication creates redundant copies of entire networks. Whole genome duplication corresponds to major transitions in evolution, such as the emergence of vertebrates.
There is direct evidence of exon shuffling mediated by mobile genetic elements in the past, and can do so experimentally in real time.
One of the most fascinating examples of retrotransposition occurring in real time is in the human brain. At least 50 % of the human genome is derived from retrotransposons, with three active families L1, Alu, and SVA that are associated with insertional mutagenesis and disease (presumably resulting from failures in or disruption of natural genetic engineering). Epigenetic and post-transcriptional suppression block retrotransposition in somatic cells, except during early embryo development, and in some malignancies. However, it is now known to occur with high frequencies in the adult brain.
Using a high-throughput retrotransposon capture and sequencing technique, an international team of researchers led by George Faulkner at University of Edinburgh in the UK identified and mapped 7 743 L1, 13 692 Alu, and 1 350 SVA somatic insertions in the hippocampus and caudate nucleus of three normal individual brains [20]. The retrotransposons mobilize to protein-coding genes differentially expressed and active in the brain. The somatic retrotranspositions far outnumber the germline retrotranspositions. Thus germline retrotranspositions accounted only for 8.4 % of Alu and 1.9 % of L1 events. While somatic insertions disproportionately affected protein-coding loci, germline insertions are rarely found in regions where they generate a deleterious phenotype. The hippocampus, a major source of adult neurogenesis, appears predisposed to somatic L1 retrotranspositions, and is consistent with the hypothesis that L1 retrotransposition is related to neural plasticity.
Intriguingly, exercise is now known to enhance cognitive functions, probably by promoting neurogenesis, and that too, is accompanied by increase in retrotransposon activity in the hippocampus of mice (see [21]).
The insertion of reversed transcribed sequences is one of the earliest examples of direct environmental feedback into germline DNA, counteracting the Central Dogma of molecular biology (see [2, 5, 22] Subverting the Genetic Text, SiS 24). Reverse transcription of spliced RNA sequences, or sequences altered by RNA editing into genetic loci can generate new genes often with chimeric coding regions due to transplicing of exons from two or more different genetic loci. RNA editing is widespread in the brain, affecting not only the function of individual genes in the short-term, but the brain’s long-term architecture that determine the states of multiple networks of genes. It has been suggested that productive epigenetic changes resulting from RNA editing are communicated back as cDNA to rewrite the genome of neurons, and that is the basis of long-term memory and high order cognition [23]. I have proposed in addition, that some rewrites can be transmitted via the germline to speed up the evolution of the brain [24] (Rewriting the Genetic Text in Human Brain Development, SiS 41). The RNA/DNA editing enzymes APOBECs have expanded in the primate lineage, and have been shown to control L1 mobility, thus, modulating somatic retrotransposition in the brain associated with adult neurogenesis and neural plasticity (see above).
New genes also arise by the reverse transcription and insertion of edited mRNAs or ncRNAs, which are now known to be responsible for a plethora of specific regulatory functions (see [16]).
Shapiro makes a convincing case that the concept of a highly specific and precisely targeted RW genome is much more compatible with the discoveries of molecule genetics than the obsolete concept of a read-only memory (ROM) subject to random accidental change. The classical concepts of physiological regulation can be applied to the control of the natural genetic engineering operators that alter DNA sequences and genome structure in non-random and controlled ways. The possibility of large coordinated changes in times of crisis can greatly speed up evolution.
Shapiro’s description on natural genetic engineering applies completely to my concept of natural genetic modification [17] that recognizes the continuity between the ‘epigenetic’ and ‘genetic’ processes of the fluid adaptable genome, the whole being in constant interrelationship with the environment in a complex network of circular causation [2, 4, 5]. Circular, nonlinear causation is responsible for the homeostasis or homeorhesis (dynamic developmental stability) of the living system at all scales, from the molecular to the ecological and social, and ultimately based on the circular thermodynamics of organisms articulated in detail elsewhere [13]. Such a nonlinear system is also predisposed to abrupt, developmental changes in high proportions of a population facing environmental challenges as the earth evolves. These nonlinear organized developmental changes are the stuff of macroevolution, the origin of major taxa defined by morphogenetic/developmental innovations [25], which are well-known to be decoupled from microevolution as defined by the change in base sequences of genes. Shapiro and others, notably John Mattick at Garvan Institute of Medical Research in Sydney Australia, have shown how step increases in non-coding DNA, along with increase in regulatory ncRNAs, transpositions and other natural genetic modification are, if not at the root of major evolutionary novelty and developmental complexity, are certainly closely correlated with it (see [26] Non-Coding RNA & Evolution of Complexity, SiS 63). What is still missing from this account is how the increase in layers of natural genetic modification translates into morphogenetic/developmental complexity, and indeed, cognitive capability.
Natural genetic modification has large implications on the safety of artificial genetic modification in particular and on the appropriate interventions for health, education, and social wellbeing in general, neither of which Shapiro has commented on.
The precision and complexity of natural genetic engineering/ modification makes clear that genetically modified organisms (GMOs) created by the crude methods generally used until very recently can only be highly unsafe [2]. Artificial genetic modification invariably interferes with natural genetic modification. In fact, it depends on disrupting and overriding the cell’s own precisely regulated natural genetic modification, which explains its total lack of precision, with many uncontrollable and unpredictable effects. Furthermore, in order to override the natural system, the synthetic genetically modified DNA molecules are forced into the cell in large numbers with stressful methods such as gene guns or electric shocks, carried by vectors designed to invade genomes, and transgenes are equipped with aggressive promoters often from viruses in order to force gene expression in the cells [17]. These stresses (as Shapiro points out) are well-known to mobilize endogenous mobile genetic elements that scramble and destabilize genomes; and account for the instability of transgenic lines, both in silencing transgenes and loss of transgenes that may have undergone unintended horizontal transfer into the genomes of organisms interacting with the GMOs.
Horizontal gene transfer is part and parcel of natural genetic engineering. GMOs released into the environment can spread antibiotic resistance marker genes as well as other bacterial and viral genes and genetic elements by horizontal gene transfer, spreading antibiotic resistance and creating new pathogens. There is already evidence for such horizontal gene transfer, and new findings suggests that even very short and damaged pieces of DNA can be taken up and incorporated by bacteria (see [27] Horizontal Spread of GM DNA Widespread, SiS 63). Insertion of transgenic DNA into genomes can cause insertion mutagenesis - including those involved in cancer development - and trigger genome instability. That is why I don’t believe GMOs can ever be safe (see [28] Why GMOs Can Never be Safe, SiS 60). I have spelt out the implications in an article entitled “The new genetics and natural versus artificial genetic modification” [17], though my article has not done sufficient justice to the exquisite precision of natural genetic modification so well described by Shapiro.
It is also on account of natural genetic modification that artificial genetic modification fails. No ecologically beneficial or complex traits have ever been successfully produced by artificial genetic modification, and even the single traits become unstable especially under environmental stress.
For the same reason, all attempts at identifying markers for common diseases or intelligence are bound to fail as they have so far failed (see [21]); while environmental epigenetic changes perpetrated on the genome are remarkably predictable.
As regards implications for social policies, we already have a great deal of knowledge on how social deprivation, psychological stress, and environmental toxins can have dire effects on us and our still unborn children and grandchildren (see [29-30] (Epigenetic Inheritance - What Genes Remember, and Epigenetic Toxicology, SiS 41), while social enrichment, caring environments and cognitive and physical exercises, and stress reducing mind-body techniques can have beneficial effects on infants, children and adults alike (see [29, 31, 32] (Caring Mothers Strike Fatal Blow against Genetic Determinism, SiS 41; How Mind Changes Genes through Meditation, SiS 63). The implications on the appropriate interventions for health, education and social well-being are clear. Hardly anyone can take comfort in the fact that some adverse environments can be worse for some people than others because we have different genes (even if such genes are identifiable). On the other hand, no one could possibly argue against the provision of optimum environmental conditions and interventions so that in the ideal, no individual’s genes or genomes would be adversely affected.
A pioneer of modern genetics Joshua Lederberg (1925-2008) invented the term euphenics [33], practices intended to improve phenotypes as opposed to eugenics, practices intended to improve genotypes. He was remarkably prescient. In the light of the fluid genome, optimising the environment for euphenics will automatically guarantee the good genes desired in eugenics, on account of the circular causation in the fluid genome. For the same reasons, no amount of eugenics or good genes will protect you from a hostile adverse environment, gene therapy and genetic modification notwithstanding.
Is euphenics so idealistic that it is just a fantasy? Not at all! They are the things most if not all people have always wanted: social equality, non-stressful work places, creative collaborative atmosphere at schools and universities as well as in society, good food produced ecologically while safeguarding natural biodiversity, renewable energies and a circular non-polluting green economy (see [34] Living, Green and Circular, SiS 53) just around the corner.
Article first published 02/06/14
Comments are now closed for this article
There are 3 comments on this article.
Christl Meyer Comment left 3rd June 2014 20:08:35
We do need respect instead of power over the living creatures! Evolution needs an open mind and discussion: http://www.christl-meyer-science.net/en/
Rory Short Comment left 3rd June 2014 01:01:32
What is needed is, what I think of as, the open-mindedness of the general supporter of i-sis, becoming more widespread. And I think it is despite the efforts of those driven by financial greed to block it.
David Collier Comment left 23rd June 2014 15:03:13
i have suspected for some time that viruses or at least fragments thereof are involved in human psychology e.g.when a belief is changed a 'viral' fragment is associated with it and when freed forms a coryza or similar.