Life after the Central Dogma
The biotech industry was launched on the scientific myth that organisms are hardwired in their genes, a myth thoroughly exploded by scientific findings accumulating since the mid 1970s and especially so since genome sequences have been accumulating (see Living with the Fluid Genome, by Mae-Wan Ho ).
We bring you the latest surprises that tell you why our health and environmental policies based on genetic engineering and genomics are completely misguided; and more importantly, why the new genetics demands a thoroughly ecological approach.
Dr. Mae-Wan Ho exposes the hidden intrigues in the vast RNA underworld where layers of interference and machinations subvert the chain of command from DNA to RNA to protein.
According to the Central Dogma, DNA, the genetic text, is read out into RNA and RNA is translated into protein. RNA is rather like the scribe copying and translating the sacred text to direct the faithful.
But geneticists are now uncovering a vast underworld of heresy to the Central Dogma where RNA agents not only decide which bits of text to copy, which copies get destroyed, which bits to delete and splice together, which copies to be transformed into a totally different message and finally, which resulting message - that may bear little resemblance to the original text - gets translated into protein. RNAs even get to decide which parts of the sacred text to rewrite or corrupt.
The whole RNA underworld also resembles an enormous espionage network in which genetic information is stolen, or gets re-routed as it is transmitted, or transformed, corrupted, destroyed, and in some cases, returned to the source file in a totally different form.
And this underworld is big, really big. The protein-coding sequence is only about 1.5% of the human genome. Yet, around 97 - 98% of the transcriptional readout of the human genome is non-protein-coding RNA. This estimate is based on the fact that intronic RNA makes up 95% of the primary protein-coding transcripts on average, and there are large numbers of non-coding RNA transcripts which may represent at least half of all transcripts. Most of the miRNAs (microRNA, see below), for example, are derived from (intergenic) regions between genes; and almost half of all transcripts from the mouse genome are non-coding RNAs. A similar estimate applies to the human genome [1].
The inescapable conclusion is that the job of mediating between DNA and protein is really the centre stage of molecular life. And who gives orders to the multitudes of RNA agents? In a sense it is everyone and no one, because the system works by perfect intercommunication. It is not the DNA, but rather, the particular environment in which the RNA agents find themselves.
For the organism (organization) to survive, it needs to turnover the DNA text continuously, adapting to the realities of its environment. In the process, it keeps certain texts invariant (see "Are ultra-conserved elements indispensable?" this series), while changing others rapidly in non-random ways (see "To mutate or not to mutate", this series). It also needs to keep referring to texts that are relevant, modifying it, or updating the interpretation in keeping with the times (see "Keeping in concert" this series).
RNA interference (RNAi) was first discovered in the nematode worm, C. elegans in the 1990s. Researchers noticed that injecting either sense RNA (the sequence that gets read and translated into protein) or antisense RNA (the complementary sequence, which does not code for protein) into the worm led to specific silencing of the gene involved. It was later found that the phenomenon was actually caused by double-stranded RNA (dsRNA) contaminating the sense or antisense RNA. RNAi now refers to all gene-silencing induced by dsRNA.
These include a host of other phenomena discovered at around the same time [2, 3]. For example, a gene could be silenced, or ‘co-suppressed’, simply by introducing an extra copy into the genome as a transgene, and transgenes themselves may be silenced either at or after transcription. The coat protein gene of a virus transferred into a plant may protect the plant from the virus, by silencing the virus’ genes.
All these phenomena are interlinked through special pathways of RNA processing that are only just being defined (see Fig. 1). Abnormal single stranded RNA (ssRNA) is turned into a double stranded RNA (dsRNA) by an RNA-dependent RNA polymerase enzyme (RDRP). The dsRNA is then chopped up into small pieces or microRNA (miRNA) by the enzyme Dicer. The same enzyme also processes certain hairpin RNA (hpRNA) and related pre-microRNA (pre-miRNA) into miRNA. The miRNA is further processed into single-stranded RNA that's incorporated into a multiprotein complex called RNA-induced silencing complex (RISC). At this point, the single stranded RNA fragment binds to complementary part of the messenger RNA and either causes the breakdown of the mRNA or prevents its translation into protein.
Remember that all this depends on complementary base pairing, just as in DNA, so these mechanisms could potentially exist for each and every one of the now estimated 24 500 genes in the genome.
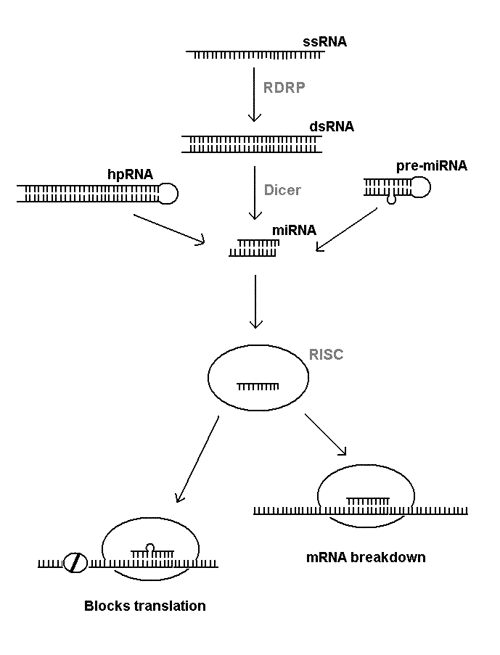
Figure 1. RNA interference pathways
It turns out that dsRNA is not only involved in signalling the breakdown or inactivation of specific mRNA to prevent the expression of the protein coded, it is also involved in triggering anti-viral response in mammals. And this is a major obstacle to achieving RNAi in mammals, which might be useful in silencing specific genes in gene therapy.
Double-stranded RNAs longer than 30 nt (nucleotide) activate an antiviral response that includes the production of interferon, resulting in the non-specific breakdown of RNA transcripts and a general shutdown of protein synthesis. In order to overcome this obstacle, synthetic 21nt miRNAs have been used. These are long enough to induce gene-specific suppression and short enough to evade host interferon response. However, recent work has shown that under certain conditions, even such small miRNAs can activate the interferon system. One activating signal for the interferon response appears to be the triphosphate group at the 5’ end of the miRNA synthesized by a phage polymerase [4]. In addition, there are other problems, such as avoiding interfering with non-target sequences [5], especially as perfect base-pairing is not required, and matches of as few as 11 consecutive nucleotides can give non-target effects.
RNA-directed DNA read-out
The dsRNA involved in RNA interference can selectively silence genes at the read-out or transcription stage [6]; dsRNA species homologous to promoters are involved in crippling the promoter by methylation (adding methyl (-CH3) groups) in the region of sequence overlap, so no transcription can occur. In other cases, a dsRNA resulting from a bi-directional transcription of a repeat element leads to methylation of a nearby histone protein H3 in chromatin, which, too, results in gene silencing.
Transcriptional gene silencing can potentially be initiated by the dsRNA formed from pairs of transcriptional units arranged in a tail-to tail orientation (sense antisense transcription units, SATs). In humans, SATs account for most overlapping transcriptional units (70%). A recent survey estimated that there are 1 600 human SATs (or 3 200 transcription units). When both transcriptional units are active, formation of dsRNA occurs by default, leading to modification of the histone protein and gene silencing. This mechanism is involved in imprinting: the marking of genes in chromosomes to determine whether they are expressed in cell clones. Expression of the gene only occurs when the antisense promoter is methylated and inactive.
Recently, a new kind of trans-acting (acting across to different parts of the genome) RNA was identified in mouse [7]. B2 RNA originates from a short interspersed repetitive element (SINE) repeated more than 105 copies in the genome of multicellular plants and animals. They were previously thought to be molecular parasites with no function. However, the level of B2 and related RNAs have been found to increase up to 100-fold in response to environmental stresses such as heat shock. And B2 RNA is required for the concomitant inhibition of RNA polymerase II during heat shock, by interacting directly with the enzyme, preventing it from working. RNA polymerase II is involved in the transcription of all protein-coding RNA. So an inhibition of RNA polymerase II will decrease the synthesis of many proteins.
A special kind of RNA directed DNA read-out is accomplished via RNA ‘riboswitches’ to switch genes off in response to the concentration of a metabolite in the cell, without the need for a protein repressor (see Box).
A new molecular switch involves an RNA molecule with enzyme activity, a ribozyme, which can self-destruct by self-cleavage [8]. This self-cleavage is accelerated 1 000 fold in the presence of a small sugar molecule, glucosamine-6-phosphate, which is generated by the enzyme protein encoded by a portion of the mRNA downstream from the ribozyme sequence.
So, this simple gene regulatory circuit involves the mRNA being translated into the enzyme, which makes the product, glucosamine-6-phospate. As the product accumulates, it binds to the special catalytic element in the mRNA, causing it to self-destruct. The region of the mRNA that can confer this regulatory activity is roughly 75 nucleotides long. When placed upstream of an un-related reporter gene, it also shuts down its expression, showing that this active RNA element is transplantable.
A particular group of ribozymes forms a pocket that binds guanosine monophosate, one of the four building blocks of RNA. A specific region of the RNA from the Human Immunodeficiency Virus (HIV) binds a derivative of the amino acid arginine. Short (<100 nucleotide) RNA aptamers (DNA or RNA molecules that bind other molecules) have been identified that specifically bind everything, from hydrophobic (water-hating) amino acids to small organic molecules and metal ions. An RNA aptamer can even distinguish the plant alkaloid theophylline from the closely related molecule caffeine.
Aptamers found within some natural mRNAs bind small molecules as part of their gene-regulatory feedback circuits. In the E. coli bacterium, coenzyme B12 binds directly to, and thereby represses translation of, the mRNA coding for the protein that transports its precursor, cobalamin. In Bacillus species, the synthesis of thiamine and riboflavin involves discrete genetic units or operons, controlled by direct binding of thiamine pyrophospate and flavin mononucleotide to leader sequences of the corresponding mRNAs, resulting in the premature termination of transcription.
Several research groups had previously engineered artificial riboswitches that accomplish exactly the same task, that is, induce ribozyme-mediated cleavage of the RNA on binding small molecules, before these were discovered in nature.
It is estimated that 64% of the genes in the human genome is interrupted [9]; i.e., the coding regions exist in short stretches (exons) interrupted by long non-coding stretches (introns). After the entire sequence is transcribed into RNA, the non-coding stretches are spliced out, leaving the coding sequence. However, different exons can be spliced together, and the borders between the exons and introns can themselves be shifted. Alternative splicing multiplies the number of different proteins that can be obtained from a single gene. This is a case of extensive cutting and pasting of the genetic text to suit the occasion.
The fruitfly gene Dscam (homologue of the Down syndrome cell adhesion molecule) codes for a cell-surface protein essential for the development of the fruitfly's brain. It has so many exons that a total of 38 016 possible alternative splice forms could be generated. Geneticists from the Whitehead Institute for Biomedical Research, Cambridge, Massachusetts in the United States analysed the splice forms expressed by different cell types and by individual cells, and found that the choice of splice variants is regulated both spatially and temporally [10].
Different subtypes of photoreceptor cells express broad yet distinctive spectra of Dscam splice forms. Individual photoreceptor cells express about 14-50 splice forms chosen from the spectrum of thousands distinctive of its cell type. Thus, the repertoire of each cell is different from those of its neighbours.
The complexity does not end there. Not only are different splice variants obtained from the same primary transcript, trans-splicing between different primary transcripts can also take place [11], multiplying the combinatorial possibilities of proteins available.
There's increasing evidence that genomic variants in both coding and non-coding sequences in genes can have unexpected deleterious effects on the splicing of gene transcripts [12]. Even synonymous base substitutions (those that do not change the amino acid sequence of the encoded protein) and sequence changes within the introns can affect splicing and cause diseases.
Some nucleotides are deleted during splicing and others changed by editing. Around 41 to 60% of mouse multi-exon genes generate alternatively spliced transcripts, the frequency of edited transcripts is unknown. These processes generate new sequences not found in the gene. Trypanosomes show the importance of RNA rewriting. Their survival depends on editing defective mitochondrial transcripts using trans-encoded RNA sequences to guide insertion and deletion of uridine bases. The rewriting of RNA restores the correct reading frame, allowing the production of functional gene products. RNA guides are also used to direct rewriting of RNA during editing and splicing of pre-mRNA. In some cases, editing creates splice sites and in others splicing prevents editing.
Rewriting of RNA is associated with a high turnover of transcripts. Of all the RNA transcribed in the human nucleus, only about 5% enters the cytoplasm Quality control mechanisms dispose of incompletely or improperly processes messages encoding flawed proteins.
Genomes can be rewritten using reverse transcription to record elements of successful ‘ribotypes’ (combination of RNAs). Around 45% of the human genome is derived from retrotransposition. RNA-directed rewriting of DNA also has an essential role in maintaining genome stability. Telomerase is a reverse transcriptase that uses an RNA guide to rewrite the ends of chromosomes (telomeres) and prevent their loss, which is important for maintaining the stability of the genome..
In each ribotype, only specific transcripts are produced and particular mRNAs translated. These outcomes are achieved by ‘coRNAs’ that coordinate the action of highly conserved pathways. An RNA product from one processing event may regulate a downstream event, making the second outcome contingent on the first. For example, a miRNA encoded in an intron would only be expressed when the host gene is transcribed. CoRNA may facilitate coordination of pathways by interacting with sequence motifs shared by a number of targets.
Evolution of rule sets requires creation of new coRNAs, possibly by duplication and mutation. New coRNAS would result in assembly of new regulatory complexes on conserved DNA elements, new patterns of gene expression during development.
Both genetic modification, involving changes in DNA, and epigenetic modifications, such as DNA methylation and histone acetylation, can be inherited. For example, imprinting is determined by the parent of origin of a chromosome, which means that at some point maternal and paternal chromosomes are marked so that they can be distinguished during embryonic development. Methylation may undergo variable erasure during primordial germ cell development, producing epigenetic mosaic individuals. The persistence of such epigenetic marks is relevant to the origin of complex diseases. Here, the susceptibility of offspring to disease can depend on whether there is maternal or paternal history of disease as well as ethnicity.
Transmission of ribotypes also occurs more directly. The embryo receives RNA from the mother that is important in specifying cells fate. The foetus is also exposed to the maternal environment, which can influence the foetal phenotype. For example, pregnant female mice fed a diet rich in methyl donors have litters with fewer yellow-coloured agouti Avy offspring, reflecting enhanced silencing of the retroviral promoter in this allele (see "Diet trumping genes", SiS 20). In other cases, integration of signals received from maternal hormones may trigger epigenetic modifications that alter long-term phenotypic development by modulating RNA co-regulatory networks. Low birth weight, for example, has been shown to correlate with lifetime risk of cardiovascular disease and diabetes mellitus.
Recently, it has been demonstrated that the plasma of pregnant women contains circulating mRNA originating from the foetus [13], which is rapidly cleared after delivery. This raises the question of whether coRNAs secreted by various somatic tissues are also used to transmit information from mother to foetus, a serious case of the inheritance of acquired characteristics not coded in the genome.
Article first published 09/09/04
Comments are now closed for this article