How quantum coherent water in the liquid crystalline cell is really responsible for the membrane potential that determines its vital states, and is crucial for energy and signal transduction Dr. Mae-Wan Ho
A new cell biology is taking shape beyond the clutter of molecular nuts and bolts that has been preventing us from seeing the wood for the trees. To clear the path, we must begin by sweeping away some deeply entrenched conventional wisdom about the membrane potential. The membrane potential has been gaining a lot of attention lately on account of its central involvement in determining the vital states of cells and tissues (see [1] Membrane Potential Rules, SiS 52); but few really understand what it is.
All cells are enclosed in a plasma membrane, and an electrical potential difference can be measured by inserting a microelectrode through the membrane into the cell or with voltage sensitive dyes. A cell at rest has an average membrane potential of -50 mV, but this can change a lot when the cell is active. As we have seen [1], changes in membrane potential determine the cell’s vital states, from the specification of body axis during development and regeneration, to cell proliferation, differentiation, and cancer. But where does the membrane potential comes from?
A typical account goes like this [2]: “The membrane potential arises from the net actions of ion channels and ion pumps (such as the sodium–potassium ATPase) embedded in the membrane, which produce different concentrations of ions (and therefore electrical charge) on the intracellular and extracellular sides of the membrane. The ion channels, when active, partially discharge the membrane potential, while the ion pumps restore and maintain it.”
That account leaves a lot unexplained. What exactly determines which ions end up inside or outside the cell? What determines the correct concentration differences inside and outside the cell?
The best (conventional) description I could find says that the membrane potential is generated by the asymmetric ion distribution across the membrane and selective ion channels, mostly K+ and Na+ channels [3]. The “simplified” explanation goes as follows.
There is a difference in the concentration of potassium across the membrane, the inside being a lot higher at about 150 mM than the 5 mM outside. The intracellular K+ is nearly balanced by the negative charges on proteins. The membrane is highly permeable to potassium, as there are many potassium channels in the cell membrane; however, the proteins cannot get out as the membrane is not permeable to proteins.
The reason the cell is slightly negative on the inside is because K+ tends to diffuse out down its concentration gradient. However, as the K+ leaves the cell, the charge balancing proteins cannot follow, so the movement of K+ out of the cell leads to the net transfer of positive charge to the outside, and consequently a small amount of unbalanced negative charge inside the cell, generating the resting membrane potential.
But why doesn’t all the K+ leave the cell? The negative charge generated inside holds them in, balancing the chemical gradient that makes them diffuse out. This is a point of equilibrium at which there is no net movement of K+ into or out of the cell.
The situation is a bit more complicated. There is a concentration gradient of Na+, much higher outside the cell at about 150 mM than the 10 mM inside. This concentration gradient and the negative membrane potential both tend to send Na+ inside the cell. The membrane also contains a small number of Na+ channels. So there is a very slow leak of sodium into the cell. Every time a Na+ leaks into the cell, a K+ ion leaves the cell to maintain electrical balance. If this were to continue, all of the K+ concentration gradient would have been dissipated. The cell’s defence against that is the Na+/K+ ATPase – the sodium potassium exchange pump, or the sodium pump – which pumps sodium out of the cell in exchange for potassium, 3 Na+ for 2 K+, for every ATP (adenosine triphosphate) hydrolysed to ADP (adenosine triphosphate) and Pi (inorganic phosphate).
The author of the above description, Sepher Eskandari, cautions [3]: “It is important to realize that the Na+/K+ ATPase is not directly responsible for the generation of the resting membrane potential …[It] is responsible for the maintenance of the membrane potential.. In fact you can poison the sodium potassium exchange pump with a chemical called ouabain.., and the cell will continue to have a resting potential for hours..”
What Eskandari has failed to explain is why K+ rather than Na+ should exist at high concentration inside the cell. He has also failed to say that the reason K+ does not leave the cell is because it is not free to do so. Instead, most, if not all of it is selectively bound to the carboxylate (COO-) groups of amino-acid side-chains on proteins. Gilbert Ling has been saying that for more than half a century, based on extensive experimental and theoretical investigations [4] (see also [5] Strong Medicine for Cell Biology, SiS 24).
When Ling proposed that practically all K+ in the cytoplasm is bound to the carboxylate groups of proteins, the cytoskeleton that pervades the cell, which we now take for granted, was not yet known (see [6] The Rainbow Ensemble, SiS 48). But there was already considerable evidence that the cytoplasm is in an organized gel-like state, rather than a disorganized thick soup of protein dissolved in water.
The evidence that K+ is bound or adsorbed, presented in detail in Ling’s book [4], includes the following [7]. There is low intracellular electrical conductance inside the cell despite the high concentration of K+, which indicates that most of the intracellular K+ is bound. Intracellular K+ mobility is strongly reduced compared to K+ outside the cell, again indicating that most of it is bound. There is a one-to-one correspondence between K+ ions and intracellular carboxylate groups. The intracellular K+ ion absorption sites were identified as the aspartate and glutamate side chains of intracellular proteins.
Among the strongest evidence that K+ is adsorbed to the cytoplasmic proteins is that the intracellular concentration of K+ remains high for hours after the cell membrane is cut or made permeable with detergent, or indeed, when the sodium potassium exchange pump is poisoned with ouabain [4].
Ling’s theory links selective K+ adsorption to a special state of intracellular proteins and especially cell water (see later). Ludwig Edelman at Saarland University in Germany, a biophysicist who took Ling’s ideas very seriously, has succeeded in obtaining the most ‘true to life’ and most beautiful electron micrographs of cells without the harsh dehydrating treatments that rob cells of their associated liquid crystalline water [8, 9] What's the Cell Really Like?, SiS 24; Why Beauty is Truth and Truth Beauty , SiS 50), and proved that K+ is selectively adsorbed to carboxylate groups of proteins in preference to Na+. That is why the cell does not need the constant action of the Na+/K+ ATPase to pump Na+ out in exchange for K+. That ‘pump’ is only ever activated to restore small deviations from a previous equilibrium; and we shall see how small and local these deviations are.
In one of many experiments [10], Edelmann incubated freeze-dried and embedded thin sections of frog muscle in solutions containing LiCl, NaCl, KCl, RbCl and CsCl (pH 7.0 Tris buffer) at concentrations of (50 mM, 50 mM, 10 mM, 10 mM and 10mM respectively), and carried out micro mass-spectroscopy to determine the amount of the different ions bound to the muscle section. He found the following order of selective ion uptake: Li+>Cs+> Rb+> K+> Na+, or Cs+> Li+ > Rb+ > K+ > Na+. Thus, K+ is invariably preferred over Na+, despite the 5-fold preponderance of Na+ over K+ in the solution.
The preferred adsorption of K+ by the cytoplasm is the equilibrium resting state, which gives rise to the potential difference between the cell and the extracellular medium, even in the absence of the cell membrane. It certainly does not require the special sodium potassium exchange pump to generate it.
Pumps and transport proteins all require ATP, which many regarded (and some still do) as the ‘energy currency’ of the cell. ATP was thought to possess ‘high energy’ phosphate bonds. But that was decisively disproved by direct measurement of the energy released in the hydrolysis of ATP in 1956 [11]. Instead, ATP functions as a major electronic modulator in the cell, as Ling has argued for nearly 60 years ago. But I am jumping ahead.
So long as the cytoplasm selectively binds K+, it will give the apparent ‘membrane potential’ measured with a microelectrode stuck into the cell. In fact, Ling’s absorption equation [4] has the same form as the usual the Goldman-Hodgkin-Katz equation for the membrane potential based on the differences in concentrations of ions outside and inside the cell [12, 13]. But contrary to the conventional assumption that the potential difference applies to the inside of the cell as a whole compared to the outside, Ling assumes that the potential difference is strictly local, applying to the bit of cytoplasm containing the tip of the microelectrode and the external medium [4]. By the same token, the change in absolute number of K+ ions that brings about a change in membrane potential locally, as in an action potential, can be extremely small, which can be readily restored to equilibrium by local pumps.
So far so good; but when proteins are tested in solution outside the cell, they almost invariably prefer to bind Na+ over K+ [6], which led to a lot of scepticism of Ling’s theory, as Ling himself recognized [4].
Both Na+ and K+ ions are hydrated (surrounded by water molecules, see [14] Dancing with Ions, SiS 49). Na+, being a smaller ion than K+, has a greater net charge on its surface than K+, and hence tends to form hydrogen bonds with water molecules. (For a primer on water and hydrogen bonds see [15] Cooperative and Coherent Water, SiS 49.) K+ being a bigger ion, has a smaller surface charge and correspondingly weaker attraction for water; so water molecules preserve their hydrogen-bonding to one another and form a cage around the K+ ion. Consequently, K+ tends to pair directly with the carboxylate group (COO-) with no water of hydration in between, whereas Na+ tends to form water-mediated pairing with the carboxylate group [7]. The two ions therefore bind very differently, with K+ ion preferring stronger acids, i.e., groups with high electron withdrawing (or low electron-donating) tendencies, whereas Na+ preferring weaker acids, i.e., groups with low electron withdrawing (or high electron donating) tendencies, which happens to be the carboxylate groups of proteins, at least outside the cell. So how can one explain the selective binding of K+ inside the cell?
Ling’s answer is that the proteins are in a very different state inside the cell, and so is the water, and the main reason is the ubiquitous presence of ATP [4].
Proteins are long linear polymers of amino-aids joined end to end in a peptide bond (-CONH-). Peptide bonds on the same chain can form hydrogen bonds with one another, giving rise to secondary structures of a-helices or b-pleated sheets. Most proteins in solution are also folded up in further globular tertiary structures. That’s the conventional textbook story.
However, that story does not apply to the cell, according to Ling. In his fully developed association-induction (AI) hypothesis, he proposed that the major components of living protoplasm – water, proteins, and K+ - exist in a closely associated, high-energy state at ‘rest’. Within the resting cell, most if not all proteins are extended so that the peptide bonds along their polypeptide backbone are free to interact with water molecules to form ‘polarized multilayers’ of aligned water molecules, while the carboxylate side chains preferentially bind K+ over Na+. Both are due to the ubiquitous presence of ATP in living cells.
In the absence of ATP, proteins tend to adopt secondary structures – a-helix, or a b-pleated sheet - as hydrogen bonds form between peptide bonds in the same chain, so they don’t interact with water (Figure 1 left). In this state, the carboxylate and amino side chains are also unavailable for binding ions, as they can pair up with each other. And the water next to the protein is not too different from the bulk phase outside the cell.
However, when ATP is bound to the ‘cardinal site’ of the protein, it withdraws electrons away from the protein chain, thereby inducing the hydrogen bonds to open up, unfolding the chain, exposing the peptide bonds on the backbone, and enabling them to interact with water to form polarized multilayers (PM) (Fig. 1 right). At the same time, the carboxylate and amino side chains are opened up to interact with the appropriate inorganic cation X+ and anion Y-. The protein ‘helper’ Z bound to the polypeptide chain is now also fully exposed. In muscle, the polypeptide chain binding ATP is myosin, and Z could well be actin. The cation X+ is K+ in preference to Na+, because ATP binding turns the carboxylate group into a strong acid that prefers K+ over Na+.
When ATP is split into ADP and Pi, and detaches from the protein, the reverse change takes place, the protein reforms its secondary structure and expels the PM water. This switching between states is the elemental ‘living machine’. It is what animates and energizes the living cell.
Notice that the state change involves a major change in the water between an ordered PM to a relatively disordered state. (Something like this could well be the basis of how actin and myosin function in muscle contraction.)
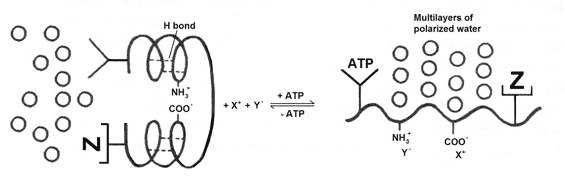
Figure 1 Gilbert Ling’s elemental living machine (redrawn from [16].
The interaction of unfolded protein chains with water is particularly significant. When protein chains are unfolded, their peptide bonds -CONH- become exposed, forming an alternating chain of negative (CO) and positive (NH) fixed charges that is very good at attracting polarized multilayers (PM) of oriented water molecules (see Figure 2). I have referred to this water as ‘liquid crystalline water’ on grounds that it forms dynamically quantum coherent units with the macromolecules ([17] The Rainbow and the Worm, The Physics of Organisms, I-SIS publication), enabling them to transfer and transform energy seamlessly with close to 100 percent efficiency. And it is this liquid crystalline water that gives the cell all its distinctive vital qualities (see [18] Life is Water's Quantum Jazz, I-SIS Lecture).
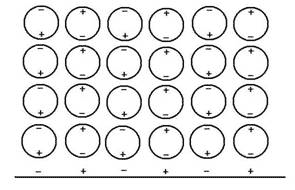
Figure 2 Polarized multilayers of water molecules form on the extended polypeptide chain
Many recent findings lend support to Ling’s hypothesis and the liquid crystalline cell, and I shall mention them in context.
PM water molecules are highly polarized and oriented. According to Ling [4], they are restricted in motion, and have shortened nuclear magnetic resonance relaxation times. (This is the basis of nuclear magnetic imaging that detects cancerous tissues by their longer relaxation times, as indicative of less structured water.) PM water does not freeze at the temperature of liquid nitrogen, and it tends to exclude solutes, which accounts for the apparent diffusion barrier for many molecules that are erroneously attributed to the cell membrane. In fact, the cell membrane offers very limited restriction to diffusion, and it is the PM water that excludes them.
PM water resembles supercooled water that has been identified in recent years as hydration water of proteins (see [19] Dancing with Macromolecules, SiS 49). A similar phase of water has been found on surfaces of hydrophilic gels, most recently by Gerald Pollack’s research team at University of Washington, Seattle in the United States (see [20] Water Forms Massive Exclusion Zones, SiS 23), which indeed excluded all solutes tested: including albumin, and pH sensitive dyes.
In addition, though not mentioned by Ling, PM is expected to be extremely good at resonant energy transfer over long distances, even better than bulk water at ambient temperatures, and to conduct positive electricity by jump conduction of protons (see [21] Positive Electricity Zaps Through Water Chains, SiS 28).
A cell with 80 percent water content would have polarized multilayers of water some 4 molecules thick that anastomose and surround the abundant cytoplasmic proteins such as those of the ubiquitous cytoskeleton. This is also precisely the thickness of the highly polarized water around proteins identified in Terahertz absorption spectroscopy within the past several years [22].
Surprisingly, an opinion review article published in 2005 stated [23]: “Recent progress in predicting protein structures has revealed an abundance of proteins that are significantly unfolded under physiological conditions. Unstructured, flexible polypeptide are likely to be functionally important and may cause local cytoplasmic regions to become gel-like.” This is another indication that Ling may well be right.
Ling sees his proposed ‘cardinal sites’ on proteins to include the ubiquitous receptor sites of cell biology, but going beyond them [4]. For example, ATP and 2,3 diphosphoglycerate (2,3 DPG), are essential for the action of haemoglobin, the iron-containing oxygen carrier protein in red blood cells. Binding of ATP and 2,3 DPG reduces the affinity of haemoglobin for oxygen, so that haemoglobin can deliver the oxygen to the lungs [24, 25]. ATP is therefore not the only cardinal adsorbent. Drugs, hormones, 2,3-DPG, Ca2+, and other potent agents at very low concentration may interact with cardinal sites to sustaining the resting living state of the protoplasm or bring about changes [16].
Thus, electronic induction is essentially the mode of action in cell [4]. The ‘cardinal adsorbents’ are electron-donating or electron-withdrawing. Induction happens via the polypeptide chain, which possesses a partially resonating structure as the peptide bond is 40 percent double bond and 60 percent single bond [26] (see Figure 3), and is therefore highly polarisable, enabling it to transfer energy and information over long distances [16].
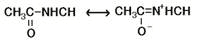
Figure 3 The resonating peptide bond
As the PM water is highly ordered and polarized if not quantum coherent [15, 17, 27] (Quantum Coherent Water and Life, SiS 51), I would expect it, too, to be equally adapt at resonant energy and information transfer, if not more so, and over the widely anastomosing networks that ultimately connect up the whole cell via the cytoskeleton.
Indeed, the major cytoskeletal proteins – actin, tubulin and intermediate filament proteins - polymerize into extended fibrous networks throughout the cell in the presence of ATP (GTP, guanosine triphosphate in the case of tubulin) [28, 29], and hence expected to support polarized multilayers of water. The cytoskeletal proteins are also all highly acidic, with glutamate and aspartate carboxylate side-chains and termini exposed and organised in clusters that are expected to show considerable preference for binding K+ over Na+ [7] in the resting polymerized state, not unlike the picture Ling had in mind. When stimulated into activity, the depolymerisation of the cytoskeletal proteins would release ATP or bound ADP and Pi, thereby bringing about a change in protein conformations that also alters the state of cell water, and with that, membrane depolarization and new chemistry due to influx of previously excluded solutes and ions [4, 7, 30] (The Importance of Cell Water, SiS 24).
Suddenly, a whole new vista has opened up. The coming decade could be the most exciting in the history of cell biology.
Article first published 12/10/11
Comments are now closed for this article
There are 1 comments on this article.
LUIS CARRAZANA Comment left 22nd September 2016 23:11:13
So, does the respiration injury giving rise to low ATP that causes Sodium and water swelling because of low ATP give rise to the Warburg effect and cancer, or is the too much Sodium in the diet that gives rise to edema causing respiration injury and Warburg's including cancer? Was Gerson's salt free diet addressing the cause of the respiration injury or a correlating fact?